Panel data from cross-sectional DAGs
Panel-data-from-cross-sectional-DAGs.RmdCreate cross-sectional DAG
Declare variables.
z <- v("z", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))
x <- v("x", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))
y <- v("y", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))Declare causal relations.

Create time series structure
Declare periods.
t1 <- v("t1", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))
t2 <- v("t2", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))
t3 <- v("t3", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))
t4 <- v("t4", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))
t5 <- v("t5", .f = d(~ rnorm(n = 10^3, mean = rsum(.x), sd = 1)))Specify persistence.

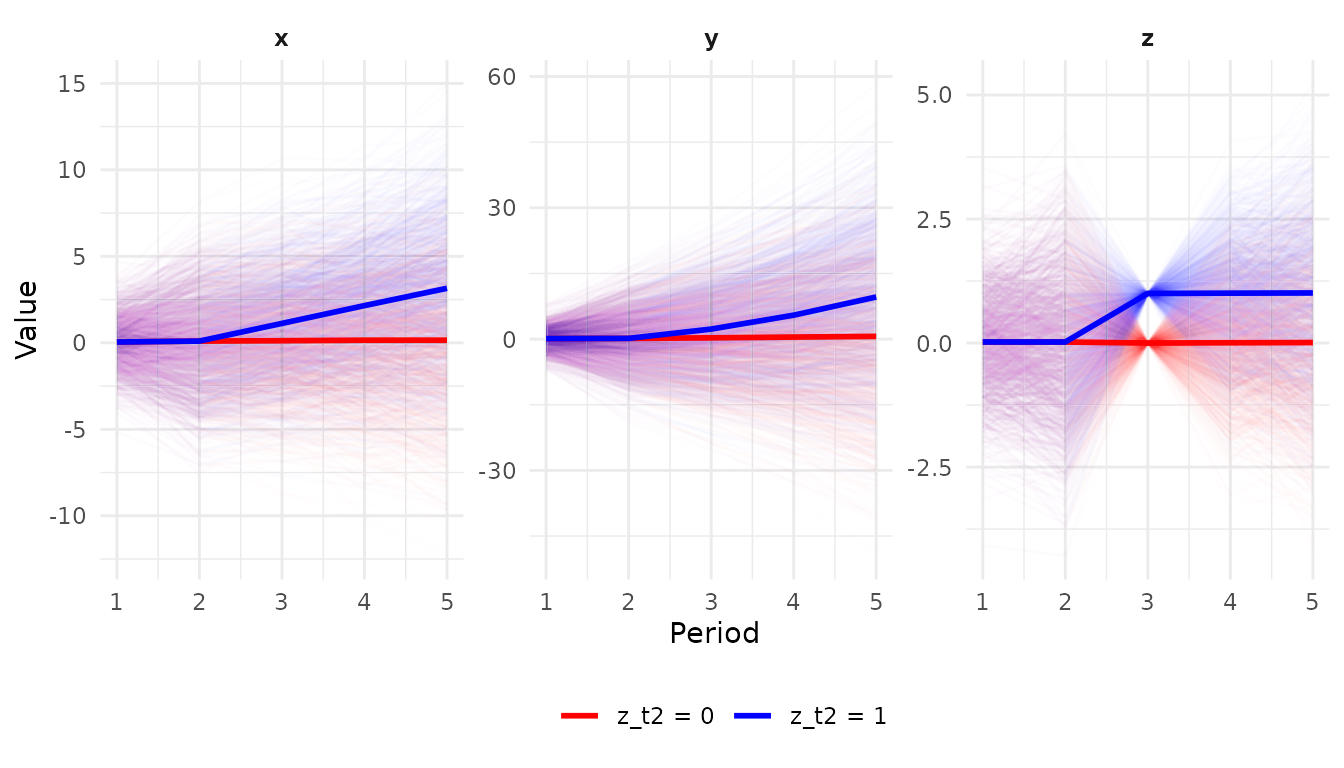
Compare intervention outcomes
Compare \(E(Y|do(z_{t3}=0)\) with \(E(Y|do(z_{t3}=1)\).
z_t2_0 <- xt %>% manipulate(z_t3 = 0) %>% simulate(label = "z_t2 = 0",seed = 2)
z_t2_1 <- xt %>% manipulate(z_t3 = 1) %>% simulate(label = "z_t2 = 1",seed = 2)
bind_rows(z_t2_0,z_t2_1) %>% panel_gather() %>% panel_plot()